Trial-aligned decoding with International Brain Lab data#
This example works through a basic pipeline for decoding the mouse’s choice from spiking activity in the International Brain Lab’s decision task, including loading the data from DANDI, processing and trial-aligning the neural activity, and fitting a logistic regression with cross-validation using scikit-learn.
The International Brain Lab’s Brain Wide Map dataset is available at Dandiset 00409. The International Brain Lab’s BWM website includes links to their preprint and additional documentation. The IBL also has an excellent decoding demonstration in the COSYNE section of their events webpage under “Tutorial 2: Advanced analysis”, along with many other relevant demos.
For a more detailed tutorial on data loading with DANDI, see the “Streaming data from DANDI” example!
Caveats: This example is meant to provide a simple starting point for working with trial-aligned data and data from the IBL, and so it does not faithfully replicate the IBL’s quality control and filtering criteria; the decoding here is also simpler than the analyses carried out in those works.
from pathlib import Path
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import pynapple as nap
import scipy.stats
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import RandomizedSearchCV
Loading data#
See also the “Streaming data from DANDI”, which includes more detail.
# These modules are used for data loading
from dandi.dandiapi import DandiAPIClient
import fsspec
from fsspec.implementations.cached import CachingFileSystem
import h5py
from pynwb import NWBHDF5IO
# The BWM dandiset:
dandiset_id = "000409"
# This is a randomly chosen recording session.
asset_path = "sub-CSH-ZAD-026/sub-CSH-ZAD-026_ses-15763234-d21e-491f-a01b-1238eb96d389_behavior+ecephys+image.nwb"
with DandiAPIClient() as client:
asset = client.get_dandiset(dandiset_id, "draft").get_asset_by_path(asset_path)
s3_url = asset.get_content_url(follow_redirects=1, strip_query=True)
fs = CachingFileSystem(
fs=fsspec.filesystem("http"),
cache_storage=str(Path("~/.caches/nwb-cache").expanduser()),
)
io = NWBHDF5IO(file=h5py.File(fs.open(s3_url, "rb")), load_namespaces=True)
nwb = nap.NWBFile(io.read())
This NWB file, loaded into a pynapple object, contains neural activity, behavioral data, and raw electrophysiological traces. They’re lazily loaded, so only what we use will be downloaded.
nwb
15763234-d21e-491f-a01b-1238eb96d389
┍━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┯━━━━━━━━━━━━━┑
│ Keys │ Type │
┝━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┿━━━━━━━━━━━━━┥
│ units │ TsGroup │
│ trials │ IntervalSet │
│ WheelMovementIntervals │ IntervalSet │
│ WheelVelocity │ Tsd │
│ WheelAcceleration │ Tsd │
│ RightSmoothedPupilDiameter │ Tsd │
│ RightRawPupilDiameter │ Tsd │
│ RightCameraMotionEnergy │ Tsd │
│ PoseEstimationRightCamera/tube_top │ TsdFrame │
│ PoseEstimationRightCamera/tube_bottom │ TsdFrame │
│ PoseEstimationRightCamera/tongue_end_r │ TsdFrame │
│ PoseEstimationRightCamera/tongue_end_l │ TsdFrame │
│ PoseEstimationRightCamera/pupil_top_r │ TsdFrame │
│ PoseEstimationRightCamera/pupil_right_r │ TsdFrame │
│ PoseEstimationRightCamera/pupil_left_r │ TsdFrame │
│ PoseEstimationRightCamera/pupil_bottom_r │ TsdFrame │
│ PoseEstimationRightCamera/paw_r │ TsdFrame │
│ PoseEstimationRightCamera/paw_l │ TsdFrame │
│ PoseEstimationRightCamera/nose_tip │ TsdFrame │
│ PoseEstimationLeftCamera/tube_top │ TsdFrame │
│ PoseEstimationLeftCamera/tube_bottom │ TsdFrame │
│ PoseEstimationLeftCamera/tongue_end_r │ TsdFrame │
│ PoseEstimationLeftCamera/tongue_end_l │ TsdFrame │
│ PoseEstimationLeftCamera/pupil_top_r │ TsdFrame │
│ PoseEstimationLeftCamera/pupil_right_r │ TsdFrame │
│ PoseEstimationLeftCamera/pupil_left_r │ TsdFrame │
│ PoseEstimationLeftCamera/pupil_bottom_r │ TsdFrame │
│ PoseEstimationLeftCamera/paw_r │ TsdFrame │
│ PoseEstimationLeftCamera/paw_l │ TsdFrame │
│ PoseEstimationLeftCamera/nose_tip │ TsdFrame │
│ tail_start │ TsdFrame │
│ LeftSmoothedPupilDiameter │ Tsd │
│ LeftRawPupilDiameter │ Tsd │
│ LeftCameraMotionEnergy │ Tsd │
│ WheelPositionSeries │ Tsd │
│ BodyCameraMotionEnergy │ Tsd │
│ OriginalVideoRightCamera │ TsdTensor │
│ OriginalVideoLeftCamera │ TsdTensor │
│ OriginalVideoBodyCamera │ TsdTensor │
│ ElectricalSeriesLf │ TsdFrame │
│ ElectricalSeriesAp │ TsdFrame │
┕━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┷━━━━━━━━━━━━━┙
The only fields we’ll use here are the neural activity (‘units’) and the trial information (‘trials’).
spikes = nwb['units']
trials = nwb['trials']
/home/runner/.local/lib/python3.12/site-packages/pynapple/core/base_class.py:50: UserWarning: Some epochs have no duration
self.time_support = IntervalSet(start=self.index[0], end=self.index[-1])
/home/runner/.local/lib/python3.12/site-packages/pynapple/core/base_class.py:52: RuntimeWarning: divide by zero encountered in scalar divide
self.rate = self.index.shape[0] / np.sum(
spikes
Index rate unit_name presence_ratio_standard_deviation contamination noise_cutoff mean_relative_depth sliding_refractory_period_violation ...
------- -------- ----------- ----------------------------------- --------------- -------------- --------------------- ------------------------------------- -----
0 15.13893 153 13.67 0.08 -0.8 20.0 1.0 ...
1 12.00565 48 20.77 0.12 79.82 20.0 1.0 ...
2 10.96317 66 24.43 0.24 309.4 20.0 1.0 ...
3 14.71296 15 15.97 0.18 224.17 20.0 1.0 ...
4 1.60872 167 6.92 0.52 71.2 20.0 0.0 ...
5 26.26409 185 1.43 0.0 16.75 20.0 0.0 ...
6 8.91826 479 0.87 0.0 16.81 40.0 0.0 ...
... ... ... ... ... ... ... ... ...
704 0.15972 644 2.73 0.0 -1.3 2540.0 0.0 ...
705 0.00049 656 0.77 0.0 -0.26 3140.0 0.0 ...
706 0.00551 698 0.59 0.0 8.5 3140.0 0.0 ...
707 0.56916 485 2.69 2.22 -0.77 3160.0 0.0 ...
708 0.00016 213 5.2 0.0 3.54 880.0 0.0 ...
709 0.08367 572 6.06 0.0 1.53 1520.0 0.0 ...
710 1.33274 328 1.16 0.0 -1.09 3140.0 0.0 ...
trials
index start end choice feedback_type reward_volume contrast_left contrast_right probability_left ...
0 36.78385458 40.007964165 -1.0 1.0 1.5 nan 1.0 0.5 ...
1 40.37566203 43.274770522 1.0 1.0 1.5 0.12 nan 0.5 ...
2 43.63566933 46.292868485 1.0 1.0 1.5 1.0 nan 0.5 ...
3 46.66926594 49.190979041 1.0 1.0 1.5 0.12 nan 0.5 ...
4 49.55207763 55.159877451 -1.0 1.0 1.5 nan 0.12 0.5 ...
5 55.57487529 60.826513102 -1.0 1.0 1.5 nan 0.25 0.5 ...
6 61.19131359 66.742723383 -1.0 1.0 1.5 nan 0.25 0.5 ...
... ... ... ... ... ... ... ... ... ...
887 4665.141205895 4674.910151903 -1.0 -1.0 0.0 0.0 nan 0.8 ...
888 4675.984954745 4690.743070591 1.0 1.0 1.5 0.06 nan 0.8 ...
889 4691.834669615 4701.794999598 -1.0 -1.0 0.0 0.12 nan 0.8 ...
890 4702.867999505 4765.977571354 0.0 -1.0 0.0 0.06 nan 0.2 ...
891 4767.122273435 4830.17841857 0.0 -1.0 0.0 nan 0.06 0.2 ...
892 4831.334918195 4834.507429035 1.0 1.0 1.5 0.12 nan 0.2 ...
893 4835.563328645 4845.873856126 -1.0 1.0 1.5 nan 1.0 0.2 ...
shape: (894, 2), time unit: sec.
Trial alignment and binning#
Here, we start by discarding some trials where a choice wasn’t made (a fuller analysis may include more criteria). Next, we create an IntervalSet aligned to the stimulus onset time within each trial, where the window around each stimulus extends back 0.5s and forward 2s.
valid_choice = trials.choice != 0
trials = trials[valid_choice]
stim_on_intervals = nap.IntervalSet(
start=trials.stim_on_time - 0.5,
end=trials.stim_on_time + 2.0,
)
Now, use build_tensor to align the neural data to these stimulus windows and bin it in 0.1s time bins.
trial_aligned_binned_spikes = nap.build_tensor(
spikes, stim_on_intervals, bin_size=0.1
)
trial_aligned_binned_spikes.shape
(711, 892, 25)
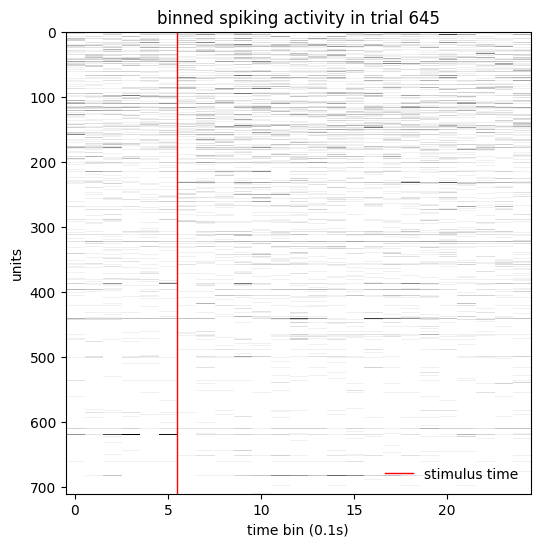
The result is n_neurons x n_trials x n_bins. Let’s visualize the neural activity in a single trial.
trial = 645
fig, ax = plt.subplots(figsize=(6, 6))
ax.imshow(
trial_aligned_binned_spikes[:, trial],
aspect='auto',
cmap=plt.cm.gist_yarg,
interpolation='none',
)
ax.axvline(5.5, lw=1, color='r', label='stimulus time')
ax.set_ylabel('units')
ax.set_xlabel('time bin (0.1s)')
ax.legend(frameon=False, loc='lower right')
ax.set_title(f'binned spiking activity in trial {trial}')
Text(0.5, 1.0, 'binned spiking activity in trial 645')
Decoding choice#
Let’s use scikit-learn to fit a logistic regression. We can use their cross-validation tools to pick the regularization parameter
# process the choice from -1, 1 to 0,1
y = (trials.choice + 1) / 2
# transpose the data to lead with trial dimension
X = trial_aligned_binned_spikes.transpose(1, 0, 2)
# standardize per neuron + trial
X = (X - X.mean(2, keepdims=True)) / X.std(2, keepdims=True).clip(min=1e-6)
# reshape to n_trials x n_features
X = X.reshape(len(X), -1)
# loguniform randomized search CV logistic regression
clf = RandomizedSearchCV(
LogisticRegression(),
param_distributions={'C': scipy.stats.loguniform(1e-3, 1e3)},
random_state=0,
)
clf.fit(X, y);
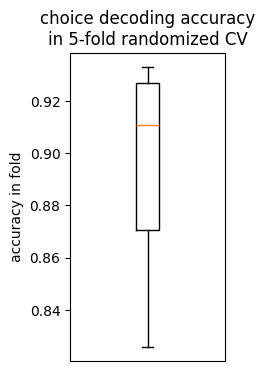
cv_df = pd.DataFrame(clf.cv_results_)
best_row = cv_df[cv_df.rank_test_score == 1]
best_row = best_row.iloc[best_row.param_C.argmin()]
plt.figure(figsize=(2, 4))
plt.boxplot(best_row[[c for c in best_row.keys() if c.startswith('split')]])
plt.ylabel('accuracy in fold')
plt.xticks([])
plt.title("choice decoding accuracy\nin 5-fold randomized CV")
Text(0.5, 1.0, 'choice decoding accuracy\nin 5-fold randomized CV')
Authors
Charlie Windolf
