Perievent#
The perievent module allows to re-center time series and timestamps data around a particular event as well as computing events (spikes) trigger average.
Peri-Event Time Histogram (PETH)#
stim = nap.Tsd(
t=np.sort(np.random.uniform(0, 1000, 50)),
d=np.random.rand(50), time_units="s"
)
ts1 = nap.Ts(t=np.sort(np.random.uniform(0, 1000, 2000)), time_units="s")
The function compute_perievent align timestamps to a particular set of timestamps.
peth = nap.compute_perievent(
timestamps=ts1,
tref=stim,
minmax=(-0.1, 0.2),
time_unit="s")
print(peth)
Index rate ref_times
------- -------- -----------
0 0 170.09
1 6.66667 182.33
2 0 186.04
3 6.66667 203.57
4 6.66667 288.25
5 0 289.45
6 3.33333 293.42
7 0 296.44
8 3.33333 320.43
9 3.33333 362.05
10 0 412.22
11 3.33333 429.92
12 3.33333 461.02
13 3.33333 465.8
14 0 487.93
15 0 495.72
16 3.33333 496.27
17 3.33333 499.53
18 0 524.52
19 0 529.15
20 0 545.65
21 6.66667 549.85
22 0 590.68
23 0 602.23
24 3.33333 645.86
25 0 659.97
26 0 664.74
27 0 665.01
28 0 671.03
29 3.33333 675.13
30 0 689.31
31 3.33333 714.83
32 6.66667 725.76
33 0 744.26
34 0 750.22
35 10 768.97
36 0 787.15
37 3.33333 788
38 0 795.03
39 3.33333 829.2
40 0 832.31
41 0 836.51
42 0 859.59
43 3.33333 911.36
44 3.33333 922.04
45 0 922.74
46 3.33333 927.94
47 3.33333 930.27
48 0 931.9
49 0 973.17
The returned object is a TsGroup. The column ref_times is a
metadata column that indicates the center timestamps.
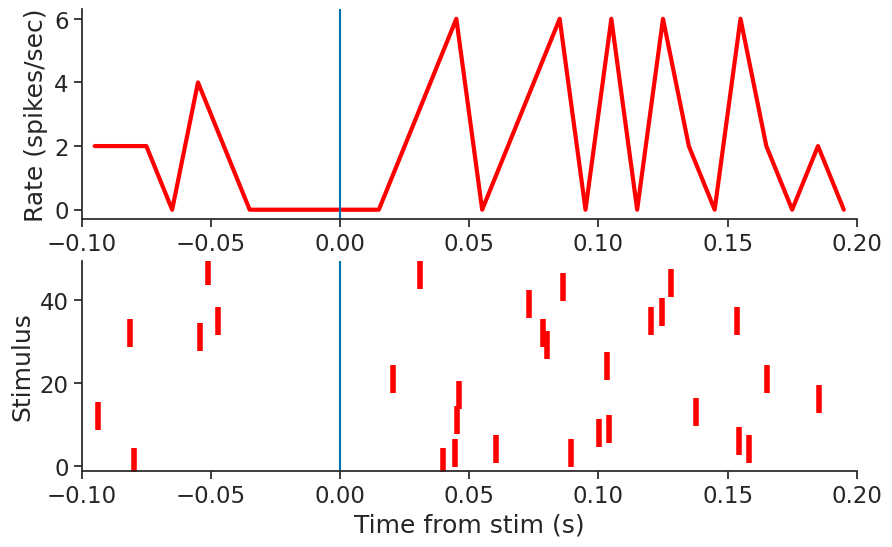
Raster plot#
It is then easy to create a raster plot around the times of the
stimulation event by calling the to_tsd function of pynapple to “flatten” the TsGroup peth.
plt.figure(figsize=(10, 6))
plt.subplot(211)
plt.plot(np.mean(peth.count(0.01), 1) / 0.01, linewidth=3, color="red")
plt.xlim(-0.1, 0.2)
plt.ylabel("Rate (spikes/sec)")
plt.axvline(0.0)
plt.subplot(212)
plt.plot(peth.to_tsd(), "|", markersize=20, color="red", mew=4)
plt.xlabel("Time from stim (s)")
plt.ylabel("Stimulus")
plt.xlim(-0.1, 0.2)
plt.axvline(0.0)
<matplotlib.lines.Line2D at 0x7fb402c2e990>
The same function can be applied to a group of neurons.
In this case, it returns a dict of TsGroup.
Event trigger average#
The function compute_event_trigger_average compute the average feature around a particular event time.
eta = nap.compute_event_trigger_average(
group=tsgroup,
feature=stim,
binsize=0.1,
windowsize=(-1, 1))
print(eta)
Time (s) 0 1 2
---------- --- --- ---
-1.0 0 0 0
-0.9 0 0 0
-0.8 0 0 0
-0.7 0 0 0
-0.6 0 0 0
-0.5 0 0 0
-0.4 0 0 0
...
0.4 0 0 0
0.5 0 0 0
0.6 0 0 0
0.7 0 0 0
0.8 0 0 0
0.9 0 0 0
1.0 0 0 0
dtype: float64, shape: (21, 3)
Peri-Event continuous time series#
The function nap.compute_perievent_continuous align a time series of any dimensions around events.
perievent = nap.compute_perievent_continuous(
timeseries=features,
tref=events,
minmax=(-1, 1))
print(perievent)
Time (s)
---------- -------------------------------
-1 [[-0.061692 ... -0.178024] ...]
0 [[-0.436613 ... -0.466229] ...]
1 [[-0.765981 ... 1.281521] ...]
dtype: float64, shape: (3, 5, 6)
The object perievent is now of shape (number of bins, (dimensions of input), number of events):
print(perievent.shape)
(3, 5, 6)
