Correlograms & ISI#
Let’s generate some data. Here we have two neurons recorded together. We can group them in a TsGroup.
ts1 = nap.Ts(t=np.sort(np.random.uniform(0, 1000, 2000)), time_units="s")
ts2 = nap.Ts(t=np.sort(np.random.uniform(0, 1000, 1000)), time_units="s")
epoch = nap.IntervalSet(start=0, end=1000, time_units="s")
ts_group = nap.TsGroup({0: ts1, 1: ts2}, time_support=epoch)
print(ts_group)
Index rate
------- ------
0 2
1 1
Autocorrelograms#
We can compute their autocorrelograms meaning the number of spikes of a neuron observed in a time windows centered around its own spikes.
For this we can use the function compute_autocorrelogram.
We need to specifiy the binsize and windowsize to bin the spike train.
autocorrs = nap.compute_autocorrelogram(
group=ts_group, binsize=100, windowsize=1000, time_units="ms", ep=epoch # ms
)
print(autocorrs)
0 1
-0.9 1.0325 0.91
-0.8 1.0425 0.94
-0.7 0.9575 1.08
-0.6 1.0075 1.06
-0.5 0.8975 1.01
-0.4 0.9050 0.98
-0.3 0.9475 1.00
-0.2 1.0750 0.97
-0.1 1.0500 1.07
0.0 0.0000 0.00
0.1 1.0500 1.07
0.2 1.0750 0.97
0.3 0.9475 1.00
0.4 0.9050 0.98
0.5 0.8975 1.01
0.6 1.0075 1.06
0.7 0.9575 1.08
0.8 1.0425 0.94
0.9 1.0325 0.91
The variable autocorrs is a pandas DataFrame with the center of the bins
for the index and each column is an autocorrelogram of one unit in the TsGroup.
Cross-correlograms#
Cross-correlograms are computed between pairs of neurons.
crosscorrs = nap.compute_crosscorrelogram(
group=ts_group, binsize=100, windowsize=1000, time_units="ms" # ms
)
print(crosscorrs)
0
1
-0.9 0.965
-0.8 1.070
-0.7 0.980
-0.6 0.925
-0.5 1.005
-0.4 1.005
-0.3 0.940
-0.2 1.000
-0.1 1.030
0.0 0.960
0.1 0.925
0.2 0.975
0.3 1.050
0.4 0.960
0.5 1.040
0.6 1.070
0.7 0.985
0.8 1.045
0.9 0.890
Column name (0, 1) is read as cross-correlogram of neuron 0 and 1 with neuron 0 being the reference time.
Event-correlograms#
Event-correlograms count the number of event in the TsGroup based on an event timestamps object.
eventcorrs = nap.compute_eventcorrelogram(
group=ts_group, event = nap.Ts(t=[0, 10, 20]), binsize=0.1, windowsize=1
)
print(eventcorrs)
0 1
-0.9 0.000000 2.777778
-0.8 1.257862 0.000000
-0.7 1.257862 5.555556
-0.6 1.257862 0.000000
-0.5 0.000000 0.000000
-0.4 1.257862 0.000000
-0.3 0.000000 0.000000
-0.2 1.257862 0.000000
-0.1 1.257862 0.000000
0.0 0.000000 2.777778
0.1 1.257862 0.000000
0.2 0.000000 2.777778
0.3 0.000000 2.777778
0.4 2.515723 0.000000
0.5 0.000000 0.000000
0.6 1.257862 0.000000
0.7 0.000000 0.000000
0.8 0.000000 2.777778
0.9 2.515723 0.000000
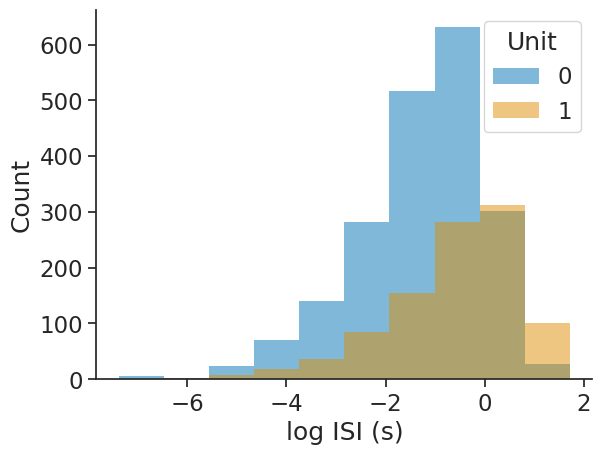
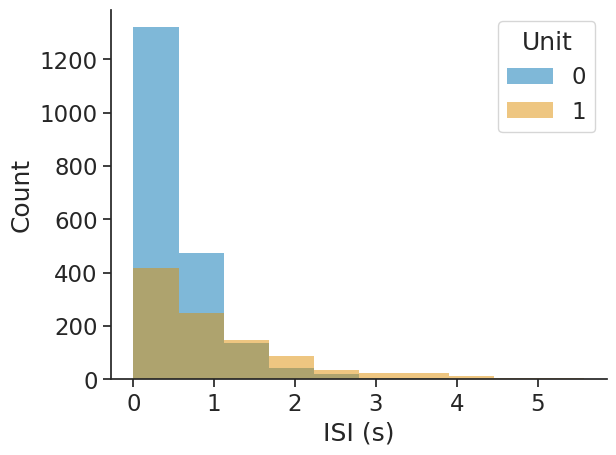
Interspike interval (ISI) distribution#
The interspike interval distribution shows how the time differences between subsequent spikes (events) are distributed.
The input can be any object with timestamps. Passing epochs restricts the computation to the given epochs.
The output will be a dataframe with the bin centres as index and containing the corresponding ISI counts per unit.
isi_distribution = nap.compute_isi_distribution(
data=ts_group, bins=10, epochs=epoch
)
print(isi_distribution)
0 1
0.279448 1321 418
0.837100 472 247
1.394752 136 146
1.952403 43 86
2.510055 19 36
3.067707 3 25
3.625359 5 23
4.183010 0 11
4.740662 0 4
5.298314 0 3
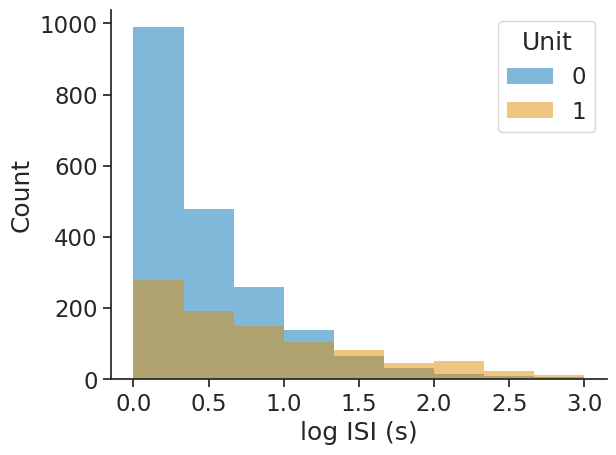
The bins argument allows for choosing either the number of bins as an integer or the bin edges as an array directly:
isi_distribution = nap.compute_isi_distribution(
data=ts_group, bins=np.linspace(0, 3, 10), epochs=epoch
)
print(isi_distribution)
0 1
0.166667 989 280
0.500000 479 193
0.833333 259 149
1.166667 138 106
1.500000 64 83
1.833333 31 46
2.166667 16 50
2.500000 10 22
2.833333 6 11
The log_scale argument allows for applying the log-transform to the ISIs:
isi_distribution = nap.compute_isi_distribution(
data=ts_group, bins=10, log_scale=True, epochs=epoch
)
print(isi_distribution)
0 1
-6.927166 6 0
-6.017077 3 2
-5.106989 23 8
-4.196900 70 19
-3.286811 140 36
-2.376723 281 85
-1.466634 516 154
-0.556546 631 282
0.353543 302 312
1.263632 27 101